Mission Statement
The major aim of our research is to create innovative and enabling technologies for biomedical and bioanalytical applications. Our research projects pursue one or more of the following goals:
- To better understand the molecular mechanisms involved in normal physiological processes and in disease
- To develop new analytical methods and instrumentation for sensitive and accurate characterization of such molecular mechanisms
- To establish and validate biomolecular patterns and signatures of disease for use in diagnostics and prognostics
- To identify and evaluate drug-candidates and affinity probes that can positively influence the outcomes of treatment
- To improve the efficiency and cost-effectiveness of our technologies through the use of innovative approaches to chemistry
Examples of ongoing projects
- Enhanced lateral-flow immunoassays
- Studying intermolecular interactions
- MicroRNA-based personalized cancer medicine
- Studying cancer, one cell at-a-time
- Better chemistry through innovative approaches
Enhanced lateral-flow immunoassays
Lateral flow immunoassay (LFIA) is a rapid, simple, and inexpensive point-of-need method. A major limitation of LFIA is a high limit of detection (LOD), which impacts its diagnostic sensitivity. To overcome this limitation, our group introduced a signal-enhancement procedure that is performed after completing LFIA and involves controllably moving biotin- and streptavidin-functionalized gold nanoparticles by electrophoresis. The nanoparticles link to immunocomplexes forming multilayer aggregates on the test strip, thus, enhancing the signal.
We lowered the LOD of hepatitis B surface antigen from approximately 8 to 0.12 ng mL−1, making it clinically acceptable. Testing 118 clinical samples for hepatitis B showed that the signal enhancement increased the diagnostic sensitivity of LFIA from 73 % to 98 % while not affecting its 95 % specificity. Electrophoresis-driven enhancement of LFIA is universal (antigen-independent), takes two minutes, and can be performed by an untrained person.

Schematic depiction of the conventional LFIA approach and electrophoretic LFIA signal amplification step.
Studying intermolecular interactions
Reversible (non-covalent) affinity interactions between molecules play an important role in every biological process, including gene expression, metabolism and progression through the cell cycle. Knowing the mechanisms that govern these binding interactions is of extreme importance to our understanding of normal cell function, disease, and drug action. At its basic level, the study of intermolecular interactions requires knowledge of their equilibrium constants (how strongly the molecules bind to each other) and their kinetics (how quickly do these binding complexes form and break apart).
Our group has invented Kinetic Capillary Electrophoresis (KCE) - a methodological platform which enables the measurement of both the equilibrium and the kinetic properties of molecular interactions in an accurate and an efficient manner. KCE is based on Capillary Electrophoresis (CE), a technique in which molecules are placed into tiny glass capillaries (as thin as a human hair) and are separated under the influence of a strong electric field.
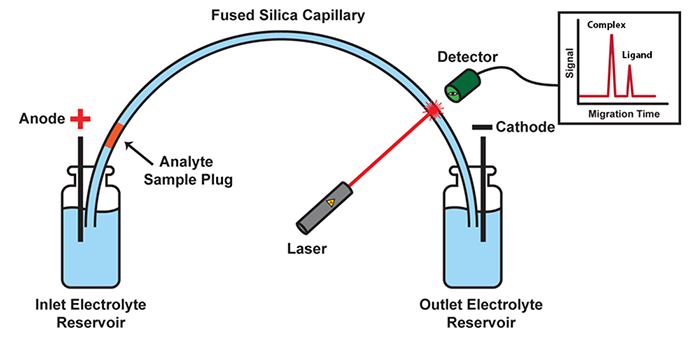
Schematic of a CE instrument: In its most basic setup, CE consists of a capillary suspended between two electrolyte-filled reservoirs, with an electrode suspended in each. Molecules move under the influence of an electric field with velocities defined by their size and charge. Molecules with differing size-to-charge ratios separate from each other, and are detected as they exit the capillary by a variety of available tools, such as laser-induced fluorescence (LIF) detection.
In KCE, molecules are allowed to interact with each other while they are being separated. The specific movement patterns of different molecules are recorded and are analyzed using sophisticated mathematical and computational techniques to extract useful information about the intermolecular interactions.
KCE has the following advantages when compared to other kinetic methods:
- High precision and accuracy
- Short analysis time
- Low sample consumption
- Relatively simple experimental setup
- High potential for automation
KCE is a highly-versatile platform and can be used for 3 different applications: (1) to measure unknown concentrations of molecules with the help of affinity probes (e.g. to perform blood tests), (2) to measure kinetic parameters of affinity interactions (e.g. for evaluation of new drugs), and (3) selection of binding ligands from highly-diverse combinatorial libraries of molecules (e.g. to discover new drugs). Due to its versatility and universality, KCE is dubbed as the Analytical Swiss Army Knife. KCE methods are being used by many laboratories all over the world. Currently, we are expanding kinetic methodology of KCE to other modes of separation, such as Kinetic Size Exclusion Chromatography (KSEC) with Mass-Spectrometry detection.
MicroRNA-based personalized cancer medicine
MicroRNAs (miRNAs) are short RNA molecules (18-25 nucleotides) that play an important role in the regulation of cellular processes. Cancer tissues have been found to have unique deregulation patterns in specific sets of miRNA, which can be used as cancer "miRNA fingerprints”. Researchers are able to distinguish between different types and stages of cancer by analyzing their miRNA fingerprint. The ability to recognize different cancer subtypes is very important for diagnostics, as this information may allow for the development of better subtype-specific treatments. A major obstacle in using miRNA in diagnostics, however, is that cells often contain it in very small quantities, making it difficult to detect.
Our group has developed a powerful detection technique called DQAMmiR (direct, quantitative analysis of multiple miRNAs). DQAMmiR is a hybridization assay that uses labeled DNA probes to recognize and bind to specific miRNA sequences. Hybridization assays require the separation of the excess, unbound DNA probe from the miRNA-DNA probe duplexes. CE is used to achieve this separation. Detection of multiple miRNA also requires the separation of the different miRNA-DNA probe duplexes. Since all miRNA are of similar length, this separation is difficult and cannot be achieved with CE alone. This was circumvented by the attachment of “drag tags” to the DNA probes which alter their size-to-charge ratio, causing a shift in their respective miRNA-DNA probe duplex migration times. DQAMmiR is able to simultaneously measure the amount of multiple miRNAs with record sensitivity and accuracy (as few as a thousand of copies of the molecule).

Schematic of DQAMmir
To further improve the sensitivity of DQAMmiR, we are developing a highly-sensitive CE instrument that takes advantage of two optical techniques: time-resolved fluorescence and confocal microscopy. These techniques are used to distinguish between the useful optical signal from miRNA molecules and the interfering signal from scattered laser light and auto-fluorescence of non-analyte materials. Our team uses advanced physics simulation software to design, optimize and build this new instrumentation. We expect that the new instrument will be able to detect as few as a single copy of a miRNA molecule.

Design and implementation of the new instrument
In the near future, DQAMmiR will be applied to real clinical samples through our working collaboration with Sunnybrook Hospital and the University of Toronto school of Medicine.
Studying cancer, one cell at-a-time
Biological tissues are made-up of many distinct types of cells that coexist alongside each other, differing in their biological function, morphology, and chemistry. Heterogeneous cell populations are generated during both normal physiological processes (e.g. embryogenesis, tissue regeneration) and during pathological processes (e.g. neurodegeneration and carcinogenesis). In fact, heterogeneity of cancer tissues is one of the major obstacles to efficient therapy, as different cell subtypes respond differently to a given treatment. Most biological studies are currently performed using "averaged" samples, where millions of cells are ground-up together, making it impossible to capture the unique properties of individual cell types. To truly understand the disease, and to create cancer subtype-specific therapy, we need to be able to study cancer tissues one cell at-a-time. Our lab has pioneered two separate technologies which allow analysis at a single-cell level: Chemical cytometry and MDRtype.
Chemical cytometry is a technology which uses microseparation methods, such as CE or nano-liquid chromatography, to study the chemical contents of individual cells. In Chemical cytometry, a single cell is injected into a capillary, lysed, stained, and subjected to capillary electrophoresis within a single experiment. Signal from various stains and their migration patterns can provide information about chemical content, metabolism and differentiation pathways of an individual cell. As an example, we have used chemical cytometry to detect asymmetry in biochemistry of two sister cancer cells, right after their division. This approach allows us to study the factors involved in differentiation of cancer cells, helping us uncover the mechanisms behind tumor tissue heterogeneity.
While chemical cytometry allows us to study the inner-workings of a cell, our MDRtype technology makes it possible to identify different cell subtypes based on their functional properties, i.e. their behavior. Multi-drug Resistance (MDR) is the ability of cancer cells to actively expel anti-cancer drugs by using membrane transport proteins. MDRtype technology uses high-content imaging instrumentation to observe a population of cells over a length of time, and track the rates of expulsion of fluorescently-labeled drugs in individual cells. We can then use computational algorithms to characterize MDR of individual cells using two kinetic parameters – strength of drug binding by transporter protein (KM) and velocity of drug transport (Vmax). We have shown that KM and Vmax can be used to distinguish between such cell populations as tumor-initiating cells and bulk tumor cells. Frequency distributions of these two parameters over a clinical sample can help us classify the the subtype of a given tumor, and help in selection of an appropriate treatment. We are planning to use MDRtype on real patient samples in collaboration with clinical scientists from Princess Margaret Hospital.

MDR capacity in different cell types - two scatterplots of KM versus Vmax uncover complex heterogeneous distribution of MDR capacity in tumor–initiating cells (TICs, right panel) compared to the uniform distribution of MDR in bulk tumor cells (BTCs, left panel).
Better chemistry through innovative approaches
Currently, most chemical processes in the pharmaceutical industry are carried out in a discontinuous fashion: reagents are mixed together, reacted, and products are purified in batches. The discontinuous nature of batch processes prevents their streamlining, as they are difficult to control and automate. A much better approach is to have the reagents mixed, reacted and products purified in a continuous-flow. This approach has a number of advantages over batch synthesis, namely: (1) increased product yield, (2) reduced costs, (3) safer operating conditions, (4) simple scale-up, (5) automated optimization and control of reaction conditions, and (6) ability to recycle unused reagents.
Our lab is working towards development of a continuous chemical synthesis system called FLOWSTREAM. The aim of FLOWSTREAM is to integrate continuous-flow purification methods based on free-flow electrophoresis (FFE) with continuous-flow synthesis to perform small scale chemical manufacturing used for the production of pharmaceuticals, research chemicals, nanomaterials, biogical polymers, etc.
It is our desire to see versatile examples of continuous and streamlined chemical production and purification become automated, straightforward and foolproof, economical, and directed towards green-chemistry processes. For this, we want to develop instrumentation and methodologies as well as applications for medicinal and process chemistry. Our research in this field includes:
- Microreactor devices for synthesis
- FFE devices for separation and purification
- On-line detection involving optical and mass spectrometrical techniques
- Automated systems for process controlling
- Electrophoretic separation of non-aqueous compounds
- Electrophoretic separation of hydrophobic compounds
- Electrophoretic separation of enantiomers
- Studying above systems and methods with computational anlaysis (using COMSOL Multiphysics®)



